DNA extraction is a fundamental process in molecular biology, genetics, and various fields of biological research. We must recognize the importance of achieving high-quality DNA extraction. It involves the isolation of DNA from cells or tissues for further analysis, such as PCR, DNA sequencing, or other molecular techniques. This article will not only cover the basics of DNA extraction, plasmid preparation and DNA isolation equipment. It will also help increase your productivity by recommending better purification techniques for different extraction needs so that you spend less time extracting DNA.
What is DNA isolation
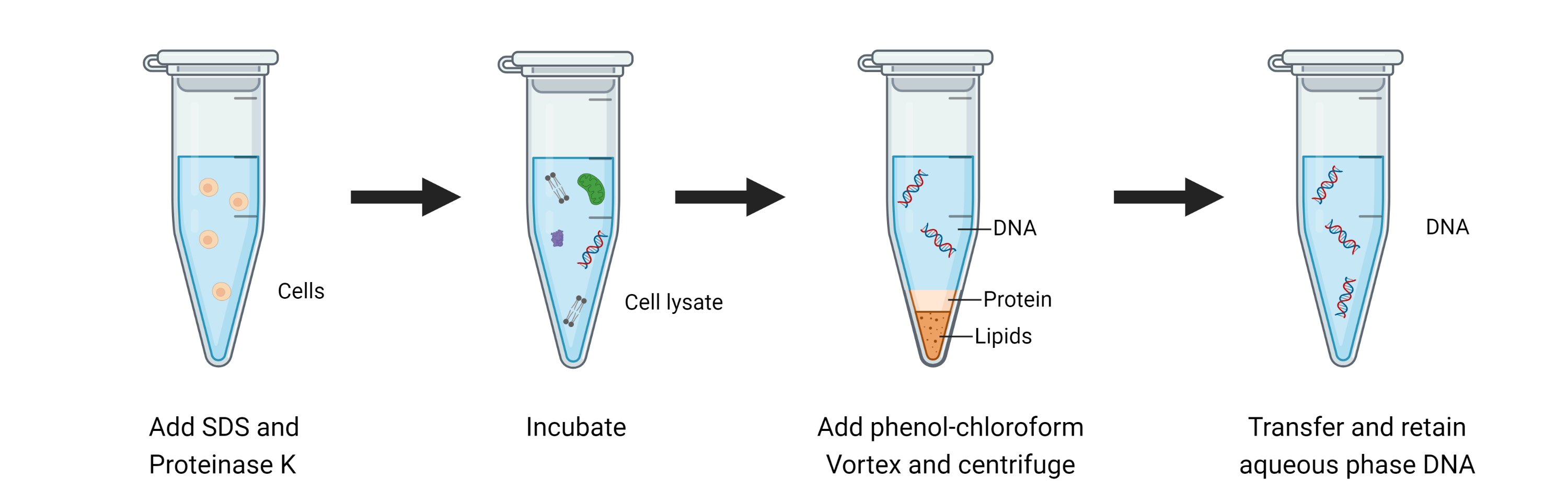
DNA isolation, also known as DNA extraction, is obtaining high-purity DNA from biological samples such as cells, tissues, or organisms. It is a fundamental step in many experimental studies, and only after DNA has been extracted with high quality can research and analysis continue. DNA extraction is significant in genetics, forensic science, genomics in PCR, DNA fingerprinting and DNA sequencing.
DNA isolation is a critical step in many areas of biology and molecular biology because the quality and purity of isolated DNA can significantly affect the success of subsequent experiments and analyses. The selection of appropriate DNA extraction methods and reagents is essential to ensure the reliability of DNA samples. It is a fundamental step in various biology and molecular research fields, including genetics, genomics, forensics, and biotechnology. You can use isolated DNA for various applications, including polymerase chain reaction (PCR), DNA sequencing, DNA fingerprinting, and genetic analysis.
Basic Steps to DNA Extraction
After collecting the biological material containing the target DNA, the cells are precipitated using a centrifuge. This will facilitate the next step of DNA extraction. The steps of DNA purification can be broadly categorized into the following five steps:
Lysis:
The first step in any nucleic acid purification reaction is to lyse the biological material containing DNA/RNA. The goal of lysis is to disrupt the cell or tissue membrane quickly, and then all components within the cell are released into the lysate, including the DNA. Four standard methods for lysing biological materials are chemical, physical, enzymatic, and combination.
Method | Feature | Experiment Type |
| Mechanical Disruption | Separation by means of a homogenizer or ultrasound is suitable for hard samples such as plant cells with strong cell walls or bacteria. Of course, for very hard samples, grinding the sample into a powder using a grinder is necessary. | Suitable for rough extractions such as environmental DNA sampling, soil DNA extraction, and DNA extraction for specialized sample types. |
| Alkaline Lysis | Alkaline Lysis is the most commonly used chemical method for lysing cells. It applies to biological materials that are easily lysed, such as tissue-cultured bacteria and fungi, as well as animal and plant cells. It primarily uses alkaline solutions (such as SDS and NaOH) to disrupt cell membranes and denature proteins. | General molecular biology experiments such as PCR, DNA sequencing and DNA fingerprinting. |
| Proteinase K Digestion | Enzymatic methods are a very gentle method of DNA cleavage and common enzyme treatments include lysozyme, lysozyme and proteinase K, lipase, collagenase and lipase. Enzymatic treatments are suitable for high throughput processing of a variety of samples, including cells and tissues, although they can be a little more expensive to use. | DNA extraction from blood or tissue samples and nucleic acid purification. Suitable for experiments requiring high DNA purity |
Protein Removal:
After Lysis, we need to separate the DNA from the solution and use a centrifuge to remove lysates. You remove proteins, sugars, or lipids from the solution by adjusting the centrifuge speed and centrifugation time. The recommended instruments to use are listed below:
| Instrument | Feature | Experiment Type |
Centrifuge
| Pros: Rapid and efficient separation of different components in solution, such as DNA, proteins and cellular debris. Suitable for a wide range of sample types and sizes. Cons: Requires specialized equipment. Usually, it requires a certain amount of sample to fill the centrifuge rotor. | Suitable for cell lysis, DNA extraction, DNA precipitation and purification steps. |
| Centrifuge Tubes | Pros: Easy to handle routine centrifugation steps. Suitable for a wide range of sample types. Cons: Manual operation, requires some lab skills。 | Suitable for centrifugation, sample partitioning, column chromatography and other separation steps. |
| Phenol/Chloroform Extraction Apparatus | This is a chemical separation that involves the use of organic solvents phenol and chloroform to separate DNA, RNA and proteins. Pros: It can effectively separate DNA, RNA and proteins. It can remove a wide range of contaminants. Cons: It uses organic compounds that are volatile and toxic. Requires careful handling. | Suitable for the isolation of DNA, RNA and proteins, especially for precise nucleic acid purification.
|
DNA Purification Columns
| Pros: Provides highly selective DNA capture and purification. Facilitates high-throughput DNA extraction. Cons: Higher cost. | Suitable for high throughput or commercial DNA extraction and purification, e.g., from blood, tissue or cell culture. |
| Magnetic Bead Separation Device | Pros: Highly selective, can be used to capture DNA or RNA. Suitable for high throughput DNA extraction. Cons: Higher price. | Suitable for high-throughput DNA or RNA extraction and nucleic acid purification. |
Each type of equipment has its unique benefits and scope of application, and the exact choice depends on the experiment's purpose, size and sample type. Some experiments may require multiple steps, combining different equipment for optimal DNA extraction and purification.
Precipitation:
A precipitation step is required to separate the DNA from the other components of the solution. It involves the addition of salt and alcohol (usually ethanol or isopropanol) to precipitate the DNA from the solution. The DNA forms visible white or turbid particles, while the contaminants remain in the liquid phase.
However, these chemicals can affect the effectiveness and purity of DNA precipitation. Therefore, DNA precipitation needs to be optimized according to experimental requirements and sample characteristics, and operating conditions need to be strictly controlled.
| Feature | Use |
Centrifuge
| Pros: Efficient Separation: No chemicals are added to ensure the quality and efficiency of the separation. Widely used: the most common tool for DNA extraction High throughput: High-speed centrifuges can handle multiple samples, making them suitable for high-throughput experiments and rapid processing. Cons: Requires specialized equipment Cannot handle large samples or sample quantities | It can effectively separate cellular residues and contaminants to ensure the purity and quality of DNA. An indispensable tool for DNA extraction and molecular biology experiments.
|
| Phenol/Chloroform Precipitation | Pros:
Highly efficient, capable of removing proteins and other impurities.
Suitable for most sample types, including cells, tissues and nucleic acid mixtures.
Cons:
It uses organic solvents and poses some safety risks to the operator.
Requires multiple steps and careful handling. | Suitable for high quality DNA and RNA purification, especially for precise nucleic acid isolation. |
| Ethanol Precipitation | Pros: Simple, economical and relatively safe. Suitable for a wide range of sample types. Cons: Less selective and may not remove all impurities. Usually, it takes longer to achieve desired precipitation. | Suitable for general DNA and RNA extraction, especially for rapid processing of large quantities of samples. |
| Isopropanol Precipitation | Pros: Simpler and safer than phenol/chloroform precipitation. Suitable for a wide range of sample types. Cons: Less selective, may not remove all impurities. Longer precipitation step. | Suitable for general DNA and RNA extraction and for laboratories requiring safer handling. |
Each sedimentation method has its characteristics and should be chosen according to the experiment's specific needs and the sample type. Centrifuges are the most efficient and provide the best sedimentation results, but they require the purchase of a machine and do not allow for high-volume processing.
Phenol/chloroform precipitation is one of the most commonly used methods because it effectively removes contaminants. However, you must handle it carefully because phenol and chloroform are toxic.
Ethanol and isopropanol precipitation methods are relatively simpler but less selective and may require more processing steps to remove impurities. Therefore, the choice of the appropriate precipitation method depends on the specific requirements of the experiment and the samples being processed.
Washing:
Washing the precipitated DNA removes residual contaminants such as salts and proteins. This is done by centrifugation, in which the DNA pellet is collected, and the washing solution is discarded.
Elution:
The purified DNA is resuspended in an elution solution, usually a low-salt buffer or deionized water. This step produces a highly purified DNA solution for downstream applications.
Solutions for different types of DNA purification
According to the following different sample purification needs, combined with our DNA extraction centrifuge or photometer and electrophoresis equipment, we can help you find the most suitable solution for your DNA purification needs!
Solid Tissue-DNA Isolation
In solid tissue genomic DNA isolation, using the appropriate type of centrifuge is important. This is because it ensures the effective separation of DNA and removal of other tissue components. When selecting a centrifuge, the required centrifugal force, centrifugation time and sample capacity need to be considered. Both high-speed and spiral centrifuges work well for DNA isolation after cleavage using Proteinase K.
Centrifugation Speed: Generally, using a centrifugation force of 14,000 to 20,000 x g (gravity) is a common range. To avoid excessive precipitation of DNA, a pre-test can be performed to determine the optimal centrifugation speed.
Centrifugation Time: Typically between a few minutes and 30 minutes. Higher centrifugation forces usually require shorter centrifugation times. In general, centrifugation should be performed for sufficient time to ensure adequate DNA extraction, but should also avoid excessive centrifugation to prevent excessive DNA precipitation.

CFG-21H
Max Speed (rpm) : 20500r/min
Max RCF (xg) : 29200
Timer : 1min-99min 59min
Max capacity : 750ml×4
Speed accuracy : ± 50r/min

CFG-19H
Max Speed (rpm) : 18500r/min
Max RCF (xg) : 23797
Timer : 1min-99min
Max capacity : 100ml×4
Speed accuracy : ± 30r/min

CFG-17H
Max Speed (rpm) : 16500r/min
Max RCF (xg) : 21500
Timer : 0min-99min 59min 59s
Max capacity : 50ml×6
Speed accuracy : ±20r/min
With a Max RCF of 29,200 x g, this centrifuge can meet the majority of DNA extraction in solid tissues. And with a large capacity of 750ml x 4, you can quickly complete a variety of high-volume separation experiments.
If you feel that the capacity of this centrifuge is too large, resulting in waste in use, we also have instruments with a Max RCF of 23797 (xg) and a capacity of 100ml×4. Of course, we also have many other high-speed centrifuges. If you are interested, you can let our engineers, according to your needs, recommend the most ideal solution!
Plant tissue-DNA extraction
The lysis of plant material is particularly difficult because it is especially difficult when dealing with tough or woody tissues. Therefore, the additional equipment required includes not only a centrifuge but also a device capable of breaking up the seeds.
CentrifugesWe usually need to use high-speed centrifuges, especially for large plant cell and tissue samples. Large plant tissues may contain more cell walls and organelles, so higher centrifugal forces are needed to separate the DNA. The high-speed centrifuge CFG-15HM is designed for manual or automated 96-well DNA purification from plant leaf and seed tissue.
The following types of centrifuges are suitable for plant DNA extraction:
CFG-15HM
Max Speed (rpm) : 15000rpm(500-15000rpm), increment: 100rpm
Max RCF (xg) : 15100×g, increment: 100×g
Timer : 1min-99min 59min
Max capacity : 0.2mL/0.5mL/1.5mL/ 2mL×12
Speed accuracy : ± 20r/min
 CFG-15H
CFG-15H
Max Speed (rpm) : 15000rpm
Max RCF (xg) : 21380×g, increment: 10×g
Timer : 30s-99min/Continuous
Max capacity : 5ml×18, 5mL culture tube×12 0.2ml/0.5ml/1.5ml/2ml×24, 0.5ml×36, PCR8×4
Speed accuracy : ± 20r/min

CFG-16H
Max Speed (rpm) : 16000r/min
Max RCF (xg) : 17800×g
Timer : 1min~99min
Max capacity : 5ml×10
Speed accuracy : ± 30r/min
Fruit tissue-DNA isolation
Most fruit DNA isolation experiments usually do not require a centrifuge. DNA is extracted from fruit samples and does not involve the separation of nuclei or cellular debris. Generally, fruit DNA extraction experiments can be performed using simple household utensils and reagents. This makes them ideal for educational and scientific activities, especially in school laboratories or home environments.
For example, the strawberry DNA extraction experiment steps:
Cut the strawberries into small pieces and place them in a plastic bag or cup.
Add salt water and detergent to the strawberries and shake gently to mix.
Strain the mixture through a plastic mesh to remove the residue and solid parts.
Collect the strawberry juice passed through the filter and pour it into a plastic funnel.
Carefully pour alcohol into the funnel to stratify it with the strawberry juice.
Observe the DNA precipitate in the alcohol layer, forming a white thread-like substance.
Carefully extract the DNA thread-like material using a plastic centrifuge tube or a small bottle.
Serum & Plasma - DNA extraction
We offer a wide selection of high-efficiency centrifuges to obtain high-quality, purified-free DNA from plasma samples. No sample pre-treatment is required. In about 60 minutes, you'll have high yields of amplifiable DNA for downstream assays, including qPCR, NGS, and digital PCR.

CFG-M12
Max Speed (rpm) : 12000r/min
Max RCF (xg) : 13500xg
Timer : 1min~99min
Max capacity : 24 pieces of capillaries
Speed accuracy : ± 30r/min
CFG-H14
Max Speed (rpm) : 200-14000rpm, increment: 10rpm
Max RCF (xg) : 18620×g
Timer : 30sec-99min/Continuous
Max capacity : 2 type
Speed accuracy : ± 20r/min

CFG-BB6
Max Speed (rpm) : 6000r/min
Max RCF (xg) : 6600×g
Timer : 1~9h 59min
Max capacity : 1000ml×6
Speed accuracy : ± 20r/min
Of course, in clinical laboratories or research, separating plasma and serum is usually done in a refrigerated centrifuge to keep the samples stable at low temperatures. The following are recommendations for the REFRIGERATED CENTRIFUGE:
CFG-T21HR
Max Speed (rpm) : 21000r/min
Max RCF (xg) : 31600×g
Timer : 0-99h 59min 59s
Max capacity : 500mL×4
Speed accuracy : ± 20r/min

CFG-T18HR
Max Speed (rpm) : 18500r/min
Max RCF (xg) : 23800×g
Timer : 1-99 min 59sec
Max capacity : 100mL×4
Speed accuracy : ± 30r/min

CFG-T15HR
Max Speed (rpm) : 15,000rpm
Max RCF (xg) : 21,380×g, step: 10×g
Timer : 30s-99 min
Max capacity : 1.5/2mL×24, 0.5mL×36, PCR8×4, 5mL×12, 5mL×18
Viruses-DNA extraction
Bacterial cells are usually small and isolation often requires high-speed centrifuges. Higher centrifugal forces are required to separate cellular debris and DNA effectively.
Gel cards - DNA extraction
Gel cards are commonly used for blood typing, disease screening, and DNA isolation and testing. In the case of DNA isolation, centrifuges are often used to separate the samples from the gel cards. The type of centrifuge suitable for gel card DNA extraction depends on the gel card used for the experiment and the desired separation conditions.
A microcentrifuge is the more common choice, as it is suitable for small sample sizes and microtubes. This type of centrifuge can usually separate samples from gel cards at high speeds for further analysis. Microcentrifuges usually have a variety of rotor and tube options for different types and sizes of gel cards.

CFG-4G
Max Speed (rpm) : 2200/2000r/min
Max RCF (xg) : 540xg
Timer : 1min~90min
No. of micro-gel (six columns) : 12cards / 24cards
Speed accuracy : ± 30r/min
CFG-15M
Max Speed (rpm) : 500~15000r/min
Max RCF (xg) : 16~15080xg
Timer : 15sec~99min59sec/∞
Rotor capacity : 12×1.5/2.0ml centrifuge tube

CFG-7M
Max Speed (rpm) : 7000r/min
Max RCF (xg) : 2680xg
Timer : Continuous operation
Tor Capacity : 0.5/1.5/2.0mL×8; 0.2mL×32 PCR tubes; 0.2mL×4 PCR 8 strips;
Consider a benchtop centrifuge or ultracentrifuge for larger samples or high-volume gel card separations. These centrifuges have higher centrifugal forces and capacities and are suitable for large-scale experiments.
Food-DNA Isolation
Food materials pose the greatest challenge for cell lysis and DNA extraction. Ultra-high or high-speed centrifuges are recommended, as they have sufficient centrifugal force and speed to separate DNA in a relatively short time while removing other components such as lipids, proteins and debris.
Plasmid DNA purification
Plasmid DNA purification is extracting plasmid DNA from microorganisms, such as bacteria or fungi, removing impurities and purifying the plasmid DNA through a series of experimental steps. A plasmid is a circular DNA molecule, usually found in microbial cells, that can carry important genetic information. Therefore, plasmid DNA purification is very important for molecular biology research and applications.
The first consideration in plasmid purification is the separation of plasmid DNA from the chromosomal DNA and cellular RNA of the host bacteria.
All Scitek plasmid isolation systems use the SDS alkaline denaturation method.
The following are general considerations for plasmid DNA extraction.
Bacterial Growth and Cultural Conditions
Successful isolation of high-quality plasmid DNA begins with culture preparation. Many factors can affect the growth of bacterial cells. Keep biomass within the acceptable range for the plasmid isolation system used, as overloading may result in poor purity and yield of plasmid DNA. The incubation time of the culture affects the yield and quality of isolated plasmid DNA. Insufficiently dense bacterial cultures produce relatively low amounts of DNA. Overgrown cultures may result in unsatisfactory yields and excessive chromosomal DNA contamination due to autolysis of the bacterial cells after reaching a stable phase. We do not recommend incubating cultures for more than 18-20 hours.
Antibiotic Selection
Most plasmids carry marker genes for specific antibiotic resistance. Only cells containing the target plasmid will proliferate by adding the selected antibiotic to the growth medium. Adding antibiotics to the desired concentration will help maximize plasmid production.
How to test DNA concentration and purity
Detecting the concentration and purity of DNA after it has been extracted is an important step in ensuring the quality of a DNA sample. There are a variety of methods that you can use for this testing, and the following are some common ones:
UV-Visible Spectrophotometer:
Spectrophotometry is one of the most commonly used methods for measuring the concentration and purity of DNA. It determines concentration by measuring the absorbance of light from a DNA solution at a specific wavelength. A typical wavelength is 260 nanometres (nm). Purity can usually be assessed by the ratio of the 260 nm wavelength to the 280 nm wavelength (A260/A280 ratio). A high-quality DNA sample typically has an A260/A280 ratio between 1.7 and 2.0.
Fluorometry
The fluorescence method uses DNA bound to a fluorescent dye to measure DNA concentration. This method is usually more sensitive than spectrophotometry and can be used for low DNA concentrations. Several commercially available DNA dyes and detection devices are available to assist in making accurate DNA concentration measurements.
Agarose Gel Electrophoresis
Agarose gel electrophoresis is a routine DNA analysis method that you can use to assess the size and purity of DNA. Although it does not accurately measure concentration, can use to detect whether DNA is contaminated or degraded.
Colourimetric methods
Colorimetric methods use a staining reagent that binds to DNA and produces a specific color change that can detect spectrophotometrically or fluorometrically. The colorimetric method provides an accurate measurement of DNA concentration.
Nanodrop
The Nanodrop is a microspectrophotometer designed to detect nucleic acid and protein concentrations. It can measure micro-level samples with high sensitivity.
Common Instruments Used in DNA Purification Experiments
The instruments and equipment typically used in DNA purification experiments depend on the specific experimental process and needs. The following are some common instruments and equipment:
Centrifuge: Centrifuges separate solid particles from liquids in a sample. In DNA purification, centrifuges separate DNA, proteins, cellular debris and other components. High-speed centrifuges, microcentrifuges, and refrigerator centrifuges are common types.
Centrifuge Accessories: Centrifuge accessories, such as rotors, tubes and holders, can be customized to suit the sample type and volume.
Thermal Cycler: Thermal cycler is used for DNA amplification techniques such as PCR (polymerase chain reaction). It can rapidly cycle at different temperatures for DNA replication and amplification.
UV-Vis Spectrophotometer: This spectrophotometer is used to measure DNA concentration and purity. It measures the absorbance of a DNA sample to determine the concentration and purity of DNA.
Electrophoresis System: An electrophoresis unit is used to separate DNA fragments to detect the size and purity of DNA. It includes an electrophoresis tank, power supply, gel and electrophoresis buffer.
Vacuum Pump and Filtration System: Used to remove waste products and solvents from a solution, usually in the solid phase extraction step of DNA purification.
PCR Instrumentation: PCR instrumentation is used to perform PCR amplification and typically includes a thermal cycler and associated PCR tubes or plates.
Vacuum Concentrator: Used to concentrate DNA or other nucleic acid samples to remove water or other solvents.
Mass Spectrometer: A mass spectrometer can be used to analyze the mass and molecular composition of a DNA or nucleic acid sample.
The choice of instrument and equipment to use depends on the DNA purification's specific process and experimental requirements. Different experiments may require different instruments to perform different steps to ensure a high-quality DNA sample is obtained.

 English
English














